Building a Simple RAG System with LlamaIndex
This code implements a basic Retrieval-Augmented Generation (RAG) system for processing and querying PDF document(s). The system uses a pipeline that encodes the documents and creates nodes. These nodes then can be used to build a vector index to retrieve relevant information.
Key Components
- PDF processing and text extraction
- Text chunking for manageable processing
- Ingestion pipeline creation using FAISS as vector store and OpenAI embeddings
- Retriever setup for querying the processed documents
Evaluation of the RAG system
Method Details
Document Preprocessing
- The PDF is loaded using SimpleDirectoryReader.
- The text is split into nodes/chunks using SentenceSplitter with specified chunk size and overlap.
Text Cleaning
A custom transformation TextCleaner is applied to clean the texts. This likely addresses specific formatting issues in the PDF.
Ingestion Pipeline Creation
- OpenAI embeddings are used to create vector representations of the text nodes.
- A FAISS vector store is created from these embeddings for efficient similarity search.
Retriever Setup
- A retriever is configured to fetch the top 2 most relevant chunks for a given query.
RAG Architecture
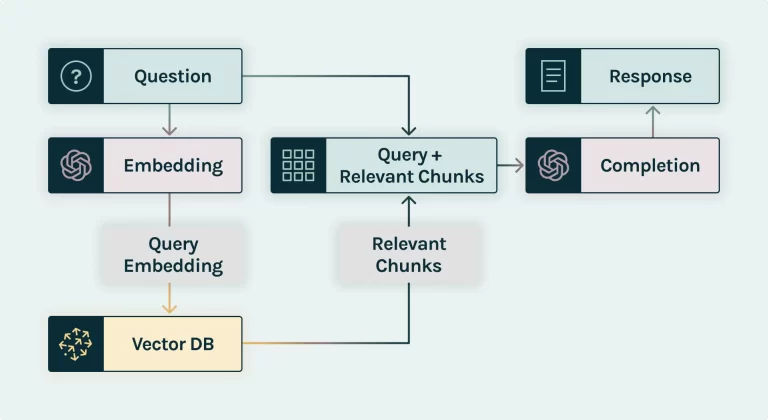
Benefits of this Approach
- Scalability: Can handle large documents by processing them in chunks.
- Flexibility: Easy to adjust parameters like chunk size and number of retrieved results.
- Efficiency: Utilizes FAISS for fast similarity search in high-dimensional spaces.
- Integration with Advanced NLP: Uses OpenAI embeddings for state-of-the-art text representation.
Code
Import libraries and environment variables
from typing import List from llama_index.core import SimpleDirectoryReader, VectorStoreIndex from llama_index.core.ingestion import IngestionPipeline from llama_index.core.schema import BaseNode, TransformComponent from llama_index.vector_stores.faiss import FaissVectorStore from llama_index.core.text_splitter import SentenceSplitter from llama_index.embeddings.openai import OpenAIEmbedding from llama_index.core import Settings import faiss import os import sys from dotenv import load_dotenv sys.path.append(os.path.abspath(os.path.join(os.getcwd(), '..'))) # Add the parent directory to the path sicnce we work with notebooks EMBED_DIMENSION = 512 # Chunk settings are way different than langchain examples # Beacuse for the chunk length langchain uses length of the string, # while llamaindex uses length of the tokens CHUNK_SIZE = 200 CHUNK_OVERLAP = 50 # Load environment variables from a .env file load_dotenv() # Set the OpenAI API key environment variable os.environ["OPENAI_API_KEY"] = os.getenv('OPENAI_API_KEY') # Set embeddig model on LlamaIndex global settings Settings.embed_model = OpenAIEmbedding(model="text-embedding-3-small", dimensions=EMBED_DIMENSION)
Read Docs
path = "../data/" node_parser = SimpleDirectoryReader(input_dir=path, required_exts=['.pdf']) documents = node_parser.load_data() print(documents[0])
Vector Store
# Create FaisVectorStore to store embeddings faiss_index = faiss.IndexFlatL2(EMBED_DIMENSION) vector_store = FaissVectorStore(faiss_index=faiss_index)
Text Cleaner Transformation
class TextCleaner(TransformComponent): """ Transformation to be used within the ingestion pipeline. Cleans clutters from texts. """ def __call__(self, nodes, **kwargs) -> List[BaseNode]: for node in nodes: node.text = node.text.replace('\t', ' ') # Replace tabs with spaces node.text = node.text.replace(' \n', ' ') # Replace paragraph seperator with spacaes return nodes
Ingestion Pipeline
text_splitter = SentenceSplitter(chunk_size=CHUNK_SIZE, chunk_overlap=CHUNK_OVERLAP) # Create a pipeline with defined document transformations and vectorstore pipeline = IngestionPipeline( transformations=[ TextCleaner(), text_splitter, ], vector_store=vector_store, )
# Run pipeline and get generated nodes from the process nodes = pipeline.run(documents=documents)
Create retriever
vector_store_index = VectorStoreIndex(nodes) retriever = vector_store_index.as_retriever(similarity_top_k=2)
Test retriever
def show_context(context): """ Display the contents of the provided context list. Args: context (list): A list of context items to be displayed. Prints each context item in the list with a heading indicating its position. """ for i, c in enumerate(context): print(f"Context {i+1}:") print(c.text) print("\n")
test_query = "What is the main cause of climate change?" context = retriever.retrieve(test_query) show_context(context)
Let's see how well does it perform:
import json from deepeval import evaluate from deepeval.metrics import GEval, FaithfulnessMetric, ContextualRelevancyMetric from deepeval.test_case import LLMTestCaseParams from evaluation.evalute_rag import create_deep_eval_test_cases # Set llm model for evaluation of the question and answers LLM_MODEL = "gpt-4o" # Define evaluation metrics correctness_metric = GEval( name="Correctness", model=LLM_MODEL, evaluation_params=[ LLMTestCaseParams.EXPECTED_OUTPUT, LLMTestCaseParams.ACTUAL_OUTPUT ], evaluation_steps=[ "Determine whether the actual output is factually correct based on the expected output." ], ) faithfulness_metric = FaithfulnessMetric( threshold=0.7, model=LLM_MODEL, include_reason=False ) relevance_metric = ContextualRelevancyMetric( threshold=1, model=LLM_MODEL, include_reason=True ) def evaluate_rag(query_engine, num_questions: int = 5) -> None: """ Evaluate the RAG system using predefined metrics. Args: query_engine: Query engine to ask questions and get answers along with retrieved context. num_questions (int): Number of questions to evaluate (default: 5). """ # Load questions and answers from JSON file q_a_file_name = "../data/q_a.json" with open(q_a_file_name, "r", encoding="utf-8") as json_file: q_a = json.load(json_file) questions = [qa["question"] for qa in q_a][:num_questions] ground_truth_answers = [qa["answer"] for qa in q_a][:num_questions] generated_answers = [] retrieved_documents = [] # Generate answers and retrieve documents for each question for question in questions: response = query_engine.query(question) context = [doc.text for doc in response.source_nodes] retrieved_documents.append(context) generated_answers.append(response.response) # Create test cases and evaluate test_cases = create_deep_eval_test_cases(questions, ground_truth_answers, generated_answers, retrieved_documents) evaluate( test_cases=test_cases, metrics=[correctness_metric, faithfulness_metric, relevance_metric] )
Evaluate results
query_engine = vector_store_index.as_query_engine(similarity_top_k=2) evaluate_rag(query_engine, num_questions=1)
This simple RAG provides a solid foundation for building more complex information retrieval and question-answering systems. By encoding document content into a searchable vector store, it enables efficient retrieval of relevant information in response to queries.
Related Post
How to Display Alert Message using Python: https://iterathon.tech//how-to-display-alert-message-using-python/
How to Impress a Girlfriend: https://iterathon.tech//how-to-impress-a-girlfriend-using-python/
Random Password Generator in Python: https://iterathon.tech//random-password-generator-in-python/

